Survival Analysis
Survival data characteristics
- in survival data, the labels are amounts of time to event
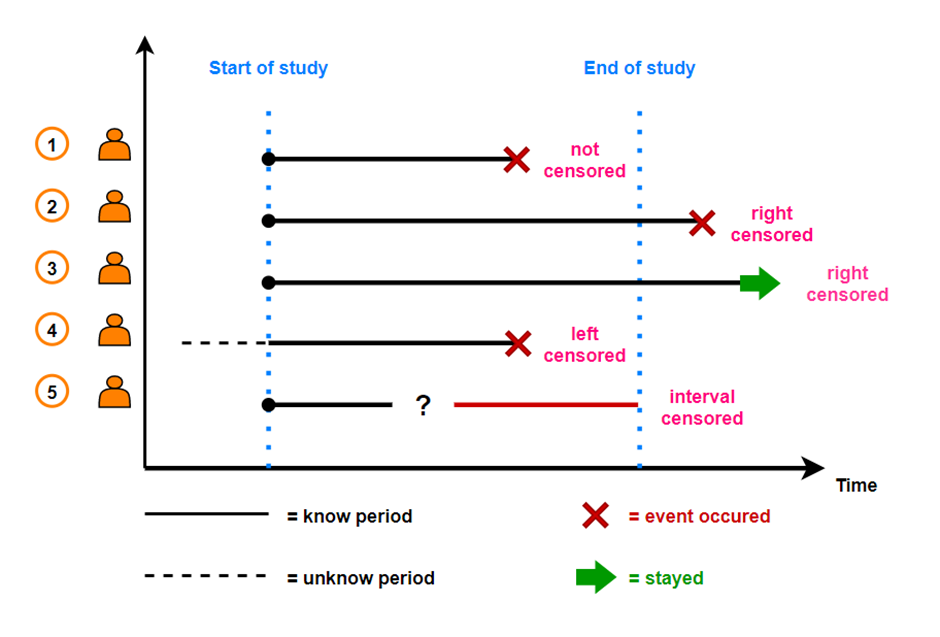
- censoring observations: no observations of events happening in the specified time period:
- end-of-study censoring (no event)
- loss-to-follow-up censoring (patients withdraw)
- left censoring = the time to events is only known to before a certain value
- right censoring = the time to events is only known to exceed a certain value (e.g. 12 months → 12m +)
(image source)
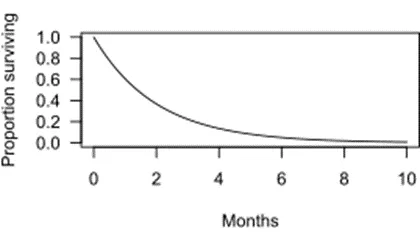
Survival function
= the probability of survival past any time t:
- always decreasing from 1 to 0: longer the t, harder to survive:
(image source) - There are parametric and non-parametric models for the survival function. Non-parametric models are mostly used in survival analysis (ref).
Parametric Survival models
- The Uniform Model (De-Moivre’s)
- Exponential Model
- The Gompertz model
- The Modified Gompertz Model (Makeham Model)
- The Weibull Model
Non-Parametric Models
Kaplan Meier Model
= the probability of survival past t months with censored observations:
where
are the events observed in the dataset is the number of deaths at time is the number of people who we know have survived up to time .
Note that Kaplan-Meier estimator estimates the survival function directly from observed data, making no assumptions about the underlying hazard function.
Hazard Function
= the instantaneous rate of death at time
= the chances of dying in a small interval of time:
- cumulative hazard
- relation between survival and hazard
Cox (Proportional Hazards) Model
= a regression model for survival data that allows us to assess the effect of covariates on survival time while making minimal assumptions about the shape of the hazard function:
where
is the hazard function for a subject with covariate values is the baseline hazard function (the hazard function when all covariates are zero) are the regression coefficients associated with covariates , respectively.
In other words, it is similar to Multiple Regression Analysis, but the difference is that the depended variable is the Hazard Function at a given time t.
When facing many features, we should consider Penalized Cox Models, and the penalization/regularization techniques are similar to linear regression:
- lasso: Regularization#^6700d3
- ridge: Regularization#^1e9ee5
- elastic net: Regularization#^de0043
Tree-structured Survival Models
- assumptions
- Cox model assumes proportional hazards, meaning the effect of covariates is constant over time.
- survival trees does not rely on the proportional hazards assumption
- non-linear relationship and high-dimensional data
- Cox model cannot handle non-linear relationship and can struggle with high-dimensional data
- survival trees can handle both
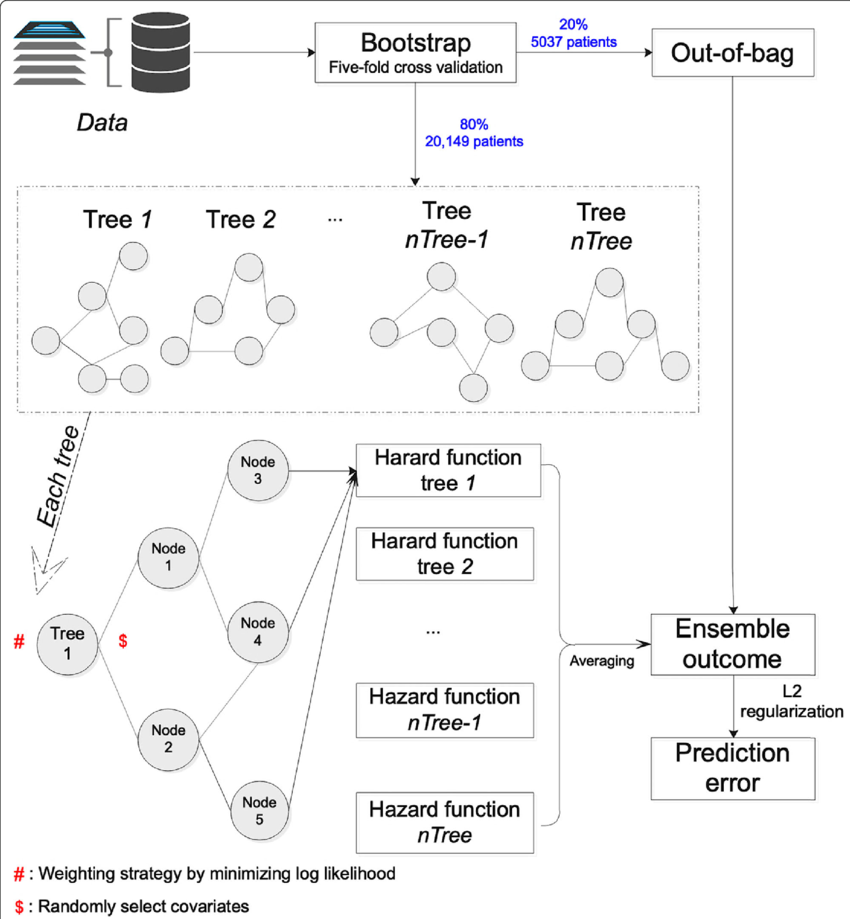
Survival trees
The single survival tree prediction for an individual is a cumulative hazard function (CHF) computed for all individuals in the same tree terminal node:
where
Survival random forest
With the CHF for each tree defined above, the entire forest the CHF averaged over all trees:
where
Deep Learning Survival Models
Example of usage
Given such a dataset:
| Subject | Time (months) | Event | Age (years) | Gender | Treatment |
|---|---|---|---|---|---|
| 1 | 10 | 1 | 55 | M | Drug A |
| 2 | 15 | 1 | 62 | F | Drug B |
| 3 | 20 | 0 | 48 | M | Drug A |
| 4 | 25 | 1 | 70 | F | Drug A |
| 5 | 30 | 0 | 58 | M | Drug B |
- To describe the survival time:
- Kaplan-Meier Estimator can be used to estimate the survival function for each treatment group
- Hazard function can be used to estimate the hazard rate changes over time for each treatment group
- To describe the effect of variables on survival:
- Cox proportional hazards model can be used to assess the effect of treatment on survival, while controlling for age and gender
Model evaluation
Graphical Evaluation
- Kaplan-Meier Curves
- Cox-Snell Residual Plot
Measure the discrimination ability of survival models
Concordance Index (C-index, or Harrell's C-index)
= quantifies the model's ability to correctly rank the relative risks or predicted survival probabilities of pairs of subjects:
- How is it computed:
- For each pair of subjects in the dataset, compare their predicted survival times or risk scores
- count concordant pair: the subject with the higher predicted survival time or lower risk score also experiences the event (e.g., death) first
- count discordant pair: If the subject with the higher predicted survival time or lower risk score does not experience the event first
- count tied pair: the predicted survival times or risk scores are equal for a pair of subjects
- C-index is then calculated as the proportion of concordant pairs among all non-tied pairs:
- The C-index ranges from 0.5 to 1.0:
- 0.5 = random choice
- 1.0 = perfect discrimination
Time-dependent Area Under the Curve (AUC)
= the AUC for different time points to assess the model's predictive accuracy over time